Multiple regulatory roles of Rad9 C-tail in DNA damage responses
Eiji Ohashi*
Department of Biology, Faculty of Science, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka, 819-0395, Japan
Abstract
DNA damage response (DDR) pathways play crucial roles in the maintenance of genome integrity, and defects in DDR proteins lead to genome instability and eventually cancer. Rad9-Hus1-Rad1 (9-1-1) is a ring-shaped heterotrimeric complex involved in multiple DDR pathways, especially the DNA damage checkpoint. Rad9 has an intrinsically disordered C-terminal region, called C-tail. The C-tail projects from the ring and has multiple phosphorylation sites and several protein-protein interaction sites, some of which are crucial for checkpoint activation. In addition, it was recently shown that C-tail binds to the 9-1-1 ring structure and is released from it upon binding to TopBP1, an activator of the DNA damage checkpoint. This review focuses on the regulatory roles of the Rad9 C-tail and discusses DNA damage checkpoint activation and the regulation of several DNA repair pathways via this region.
Text
The 9-1-1 clamp is a heterotrimeric complex loaded onto DNA by the Rad17-RFC clamp loader upon DNA damage
Multiple DNA damage response (DDR) pathways respond to DNA damage. One important mechanism involved in these pathways is the DNA damage checkpoint, which coordinates cell cycle arrest, DNA repair, and even apoptosis by phosphorylating proteins involved in these processes. The ATM (ataxia telangiectasia-mutated) and ATR (ATM- and Rad3-related) pathways are two major DNA damage checkpoint pathways in eukaryotes1. Mutations of the ATM and ATR genes are associated with the rare autosomal recessive disorder Ataxia telangiectasia and a form of Seckel syndrome, respectively. In addition, it was recently reported that a germline mutation of ATR is linked to a hereditary oropharyngeal cancer syndrome2. ATM and ATR are serine/threonine (S/T) kinases that phosphorylate a wide variety of substrates and control their functions in response to DNA damage. While ATM predominantly responds to DNA double-strand breaks (DSBs) caused by ionizing radiation (IR), for example, ATR responds to single-stranded DNA (ssDNA) generated after ultraviolet (UV) light irradiation, for example. When DNA polymerases stall at such damaged sites, and DNA helicase passes through the lesion, uncoupling of the DNA polymerase from the helicase occurs, resulting in the exposure of ssDNA3. ssDNA is also generated via several DNA repair pathways. Thus, the ATR pathway has the potential to respond to a wide variety of DNA damage. One of the key factors in the ATR pathway is the 9-1-1 checkpoint clamp.
The 9-1-1 checkpoint clamp is a heterotrimeric complex composed of Rad9, Hus1, and Rad14-6. It has a ring structure similar to PCNA (proliferating cell nuclear antigen). PCNA is a homotrimer that is loaded onto DNA by its clamp loader, RFC (replication factor C)7,8. PCNA acts as a processivity factor of DNA polymerases during DNA synthesis, encircling DNA and tethering the DNA polymerase to DNA. It also acts as a scaffold to recruit other proteins such as DNA repair proteins and chromatin-remodeling factors, depending on the context. In response to DNA damage, 9-1-1 is loaded onto DNA by another specific clamp loader, Rad17-RFC. In the Rad17-RFC complex, the largest subunit of RFC1 in the authentic RFC complex (RFC1-5) is substituted by Rad17. The loaded 9-1-1 is also thought to function as a scaffold similar to PCNA to recruit the many proteins involved in several DNA repair pathways, apoptosis, and the DNA damage checkpoint. Because genetic studies have shown that Rad9, Hus1, Rad1, and Rad17 are in the same epistasis group, 9-1-1 complex formation and its loading by Rad17-RFC are considered important for the functions of these factors4-6. The 9-1-1 complex is thought to mainly function as a sensor of the ATR-dependent checkpoint pathway9,10.
The initial step of ATR activation is the binding of RPA (replication protein A) to extended ssDNA regions after DNA damage. ATR and its binding partner, ATRIP (ATR-interacting protein), are recruited to RPA-bound ssDNA via an interaction between ATRIP and RPA. Independently, 9-1-1 is loaded by Rad17-RFC in an RPA-dependent manner. An ATR activator, TopBP1 (DNA topoisomerase II β-binding protein 1), binds to both ATR-ATRIP and 9-1-1 complexes, and both interactions are required for the full activation of ATR. Thus, 9-1-1, together with TopBP1, plays a role in the early step of DDR9,10.
Phosphorylation of S/T near the end of Rad9 C-tail facilitates ATR activation
A component of 9-1-1, Rad9, has a PCNA-like domain [~270 amino acids (aa) in human], as do Hus1 and Rad1, and an intrinsically disordered C-terminal extension (~120 aa in human), called C-tail. The C-tail is not involved in formation of the ring structure and interacts with its several binding partners including RPA and TopBP1 (Figure 1). In mammals, Rad9 C-tail has at least 10 phosphorylation sites (Table 1). Some of these sites are phosphorylated in a cell cycle- or DNA damage-dependent manner, while others are constitutively phosphorylated11,12.
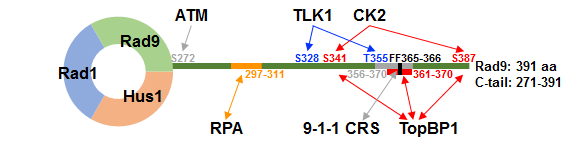
Figure 1: Rad9 C-tail has multiple phosphorylation and interaction sites.
Only the phosphorylation (single-headed arrows) and interaction (double-headed arrows) sites in human Rad9 C-tail described in this review are shown. Phosphorylation by CK2 is required for interaction with TopBP1 and for subsequent activation of DDR (red arrows), while that by TLK1 contributes to termination of DDR (blue arrows). Interaction between RPA and Rad9 is important for 9-1-1 recruitment to the site of DNA damage (an orange arrow). The function of phosphorylation by ATM and the interaction between C-tail and 9-1-1 core ring structure (CRS) are not well understood (grey arrows). Note that there are other sites that are not shown here (see also Table 1).
| Amino acid | Sequencea | Kinase | Regulation | References |
|---|---|---|---|---|
| S272 | SHSQDL | ATM, other | IR, HU, and UV-induced | 11, 12, 49 |
| S277 | LGSPER | CDK1, other | constitutive/mitotic | 11 |
| T292 | HSTPHP | CDK1 | mitotic | 11 |
| S328 | SLSPGP | CDK1, TLK1 | constitutive/mitotic/fluctuates upon IR | 11, 36, 37 |
| S336 | PKSPGP | CDK1, other | constitutive/mitotic | 11 |
| S341 | PHSEEE | CK2 | constitutive | 12, 23 |
| T355 | PGTPPP | CDK1, TLK1 | constitutive/mitotic/fluctuates upon IR | 11, 38 |
| S375 | VRSPQG | CDK1 | 12 | |
| S380 | GPSPVL | CDK1 | 12 | |
| S387 | EDSEGE | CK2 | constitutive | 11, 23, 24 |
aAmino acid sequence surrounding phosphorylated residue (underlined).
Table 1: Phosphorylation sites in human Rad9 C-tail
Phosphorylation of the DNA damage-inducible site (S272) is not important for cell survival or G2/M arrest after DNA-damaging treatments11,12. On the other hand, a constitutive phosphorylation site, S387, is important for Rad9 C-tail interaction with TopBP1 and the checkpoint activation after IR, UV exposure, or hydroxyurea (HU) treatment, a ribonucleotide reductase inhibitor11,13,14. It was also reported that the N-terminal tandem BRCT (BRCA1 C-terminal) domains of TopBP1 bind to Rad9 in a manner dependent on phosphorylation of S387 (S373 in Xenopus)13,14. Rad9 was proposed to play a role in recruitment of TopBP1 to the stalled replication fork for the activation of ATR13-15. On the other hand, several reports showed that depletion of TopBP1 reduces recruitment of 9-1-1 (or Rad9) to sites of replication stress or damage and that the interaction is dispensable for TopBP1 localization16-21. More recently, it was reported that TopBP1 binds to RPA-bound ssDNA via its BRCT2 domain22. Thus, the simple model that 9-1-1 recruits TopBP1 to stressed sites needs to be modified. 9-1-1, TopBP1, and ATR-ATRIP can be recruited to the same target, RPA-ssDNA complexes, independently of each other; however, subtle differences in conditions (e.g., cell cycle status, the amount and type of DNA damage, or the level of checkpoint activation) may influence their recruitment.
The phosphorylation of S387 is catalyzed by casein kinase 2 (CK2)23,24. CK2 also phosphorylates S341 of Rad9, and phosphorylation of both S387 and S341 is required for efficient binding to TopBP1 and subsequent activation of ATR23,25. Consistent with the constitutive phosphorylation of S341 and S387, the interaction between 9-1-1 and TopBP1 is also constitutive23. Thus, this interaction may be a prerequisite for ATR activation to ensure immediate activation of ATR in response to DNA damage. DNA-damage inducibility of ATR activation may be conferred by other factors or other interactions. Interestingly, the phosphorylation sites in yeast Rad9s [Saccharomyces cerevisiae (sc) Ddc1 and Schizosaccharomyces pombe (sp) Rad9], which are responsible for binding to TopBP1 (scDpb11 and spRad4/Cut5), are phosphorylated by yeast ATR and ATM kinases (scMec1, scTel1, spRad3, and spTel1) in a DNA damage-inducible manner26-28. In S. pombe, the interaction is also DNA damage-inducible26. During evolution, higher eukaryotes may have acquired more complex mechanisms for ATR activation. Recently, RHINO (Rad9-, Hus1-, Rad1-interacting nuclear orphan) was identified as a co-binding factor of TopBP1 and 9-1-129. RHINO is recruited to sites of DNA damage in a manner dependent on the 9-1-1 complex and is involved in Chk1 phosphorylation upon DNA damage29. RHINO is conserved among vertebrates and forms a stoichiometric complex with 9-1-130. Interestingly, loss of RHINO reduces the ATR signal upon UV irradiation without affecting the interaction between 9-1-1 and TopBP130.
While 9-1-1 is well known to be involved in ATR activation, the clear evidence that 9-1-1 contributes to ATM activation has not been provided. However, 9-1-1 was shown to participate in DSB repair mediated by homologous recombination (HR) by interacting with Rad51 recombinase31. Interestingly, tailless Rad9, which lacks C-tail, partially rescues HR defects, suggesting that the roles of 9-1-1 in the ATR checkpoint activation and HR are distinct32. Recently, Rad17 was shown to recruit the MRN (MRE11, Rad50, and Nbs1) complex to DSB sites and to participate in ATM checkpoint signal amplification33. Because the MRN complex is also involved in ATR activation18,19,34,35, crosstalk may occur between DSB repair and ATR activation.
Rad9 C-tail plays multiple roles in the DDR
The functions of 9-1-1 are regulated diversely by Rad9 C-tail (Figure 1). As described above, phosphorylation of Rad9 by CK2 is required for binding to TopBP1 and activation of ATR. In addition, Tousled-like kinase 1 (TLK1) and a splice variant of TLK1, TLK1B, phosphorylate Rad9 at S328 and T355, both of which are also target sites for CDK1 kinase, and control the recruitment and dissociation of 9-1-1 at DSB sites, thereby regulating the cell cycle checkpoint and DNA repair11,36-38. In S. pombe, Dbf4-dependent kinase phosphorylates Rad9 at S319, S320, and T321 following phosphorylation of T225 by ATR (spRad3)39. These phosphorylation events are required to release Rad9 from chromatin by decreasing its affinity for RPA and may facilitate the transition from DNA damage signaling to DNA repair.
In addition to its phosphorylation sites, C-tail has multiple protein-protein interaction sites, including those that mediate interactions with TopBP1 and RPA. C-tail also has a nuclear localization signal, which is important for nuclear targeting of Hus1 and Rad140. Interestingly, Caspase-3, a frequently activated death protease, cleaves Rad9 at multiple sites, and the cleaved N-terminal portion of Rad9 localizes to the cytosol and induces apoptosis41.
Finally, the interaction between the intrinsically disordered Rad9 C-tail and the 9-1-1 core ring structure (CRS) may also play a regulatory role in the DDR (Fig. 2). 9-1-1 binds to DNA in the absence of Rad17-RFC, and this binding is enhanced by the deletion of Rad9 C-tail42,43. Interestingly, Rad9 C-tail binds to 9-1-1 CRS, and this intramolecular interaction interferes with the binding of 9-1-1 to DNA43. The region of the C-tail necessary for binding to 9-1-1 CRS partially overlaps with that necessary for binding to TopBP1. Indeed, 9-1-1 CRS and TopBP1 bind to C-tail in a competitive manner. Interestingly, two tandem phenylalanine residues (F365 and F366) in C-tail are important for binding to both CRS and TopBP143. Two adjacent aromatic residues are found in many protein-protein interaction motifs such as the PCNA-interacting protein (PIP) motif (often called PIP-box)44, the REV1-interacting region motif45, and the Mlh1-interacting protein motif46. Aromatic residues in PIP-box fit into a large hydrophobic pocket on the front face of PCNA that is formed by residues from the interdomain connecting loop and nearby β-strands. Rad9 and Hus1 have similar structures to the hydrophobic pocket of PCNA47. Therefore, the FF sequence of Rad9 C-tail may interact with either one or more hydrophobic pockets in 9-1-1 subunits. Furthermore, when TopBP1 binds to C-tail, the C-tail will be released from the CRS (Unfolded, see Fig. 2) and the hydrophobic pocket(s) of 9-1-1 CRS will be exposed43. Many DDR proteins are reported to interact with 9-1-1; therefore, some of them harboring tandem aromatic residues may bind to the hydrophobic pocket(s) of 9-1-1 CRS in a manner similar to the binding of a PIP-box protein to PCNA. While tandem aromatic residues are part of one of the 9-1-1 CRS-binding motifs, another 9-1-1-binding motif of budding yeast Red1, a component of the synaptonemal complex, is also interesting. Eichinger and Jentsch reported that yeast Red1 has two Qxx(L/I/V/M) motifs, both of which are involved in two 9-1-1 subunit-specific interactions48. One is for binding to Hus1 (scMec3) and the other is for binding to Rad9 (scDdc1). These motifs are similar to part of the typical PIP-box [Qxx(L/I/M/V)xx(F/Y)(F/Y)]; therefore, they speculated that the hydrophobic pockets of 9-1-1 CRS and hydrophobic residues in the 9-1-1-binding-motifs of Red1 are important for their binding5,48. Thus, 9-1-1-binding proteins may have several types of motifs that are similar but not identical to the PIP-box.
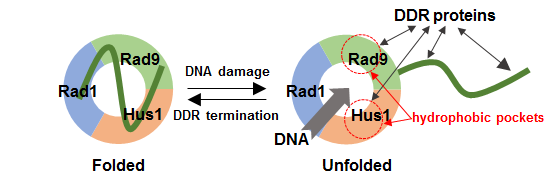
Figure 2: The intramolecular interaction of Rad9 C-tail may have a regulatory role.
In response to DNA damage, 9-1-1 is loaded onto DNA and activates DDR, interacting with several DDR proteins such as RPA and TopBP1 through Rad9 C-tail. When C-tail is unfolded upon loading, 9-1-1 can bind to DNA through its central hole, and hydrophobic pockets of 9-1-1 may be exposed and targeted by other DDR factors. The intramolecular interaction between Rad9 C-tail and 9-1-1 CRS may inhibit promiscuous protein-protein interactions after DDR termination. See the text for more details.
Future directions
9-1-1 is a PCNA-like heterotrimeric complex. Some PCNA-interacting proteins involved in DDR, including base excision repair, mismatch repair and cell cycle regulation, also interact with 9-1-15,6. Depending on the cellular context, these proteins may bind to PCNA or 9-1-1 clamps, resulting in the expression of distinct functions and different outcomes. Although the mechanism is not clear, modification(s) of Rad9 C-tail and/or intramolecular interaction between Rad9 C-tail and 9-1-1 CRS may contribute to this type of clamp switching mechanism. Rad9 C-tail may also contribute to checkpoint activation and termination via known and unknown mechanisms. While co-crystal structures of PCNA and its binding peptides have been well studied, those of 9-1-1 have not yet been reported. Thus, the binding mechanism of 9-1-1 to its partners at the atomic/molecular level is poorly understood. One of the most critical issues to be addressed is the biological significance of the binding of these DDR proteins to 9-1-1; however, this will only be forthcoming when specific mutant(s) defective only in binding to 9-1-1 are subjected to intensive study. It will also be important to define better the binding sites in 9-1-1 that mediates its binding to multiple partners as well as the binding mechanism to understand how 9-1-1 plays such a wide variety of roles in genome maintenance.
Acknowledgements
This study was supported by KAKENHI (22770172 and 16H04743) and Uehara Memorial Foundation (103-2012).
References
-
- Awasthi P, Foiani M, Kumar A. ATM and ATR signaling at a glance. J Cell Sci. 2015; 128: 4255-4262.
- Tanaka A, Weinel S, Nagy N, et al. Germline mutation in ATR in autosomal- dominant oropharyngeal cancer syndrome. Am J Hum Genet. 2012; 90: 511-517.
- Byun TS, Pacek M, Yee MC, et al. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005; 19: 1040–1052.
- Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst). 2004; 3: 1009-1014.
- Eichinger CS, Jentsch S. 9-1-1: PCNA's specialized cousin. Trends Biochem Sci. 2011; 36: 563-568.
- Broustas CG, Lieberman HB. Contributions of Rad9 to tumorigenesis. J Cell Biochem. 2012; 113: 742-751.
- Choe KN, Moldovan GL. Forging Ahead through Darkness PCNA Still the Principal Conductor at the Replication Fork. Mol Cell. 2017; 65: 380-392.
- Shiomi Y, Nishitani H. Control of Genome Integrity by RFC Complexes Conductors of PCNA Loading onto and Unloading from Chromatin during DNA Replication. Genes (Basel). 2017; 8: E52.
- Burrows AE, Elledge SJ. How ATR turns on TopBP1 goes on ATRIP with ATR. Genes Dev. 2008; 22: 1416-1421.
- Cimprich KA, Cortez D. ATR an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008; 9: 616-627.
- St Onge RP, Besley BD, Pelley JL, et al. A role for the phosphorylation of hRad9 in checkpoint signaling. J Biol Chem. 2003; 278: 26620-26628.
- Roos-Mattjus P, Hopkins KM, Oestreich AJ, et al. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J Biol Chem. 2003; 278: 24428-24437.
- Delacroix S, Wagner JM, Kobayashi M, et al. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007; 21: 1472-1477.
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007; 282: 28036-28044.
- Greer DA, Besley BD, Kennedy KB, et al. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003; 63: 4829-4835.
- Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. 2009; J Cell Biol. 184: 793-804.
- Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010; 21: 926-935.
- Lee J, Dunphy WG. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013; 24: 1343-1353.
- Duursma AM, Driscoll R, Elias JE, et al. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013; 50: 116-122.
- Gong Z, Kim JE, Leung CC, et al. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010; 37: 438-446.
- Ohashi E, Takeishi Y, Ueda S, et al. Interaction between Rad9-Hus1-Rad1 and TopBP1 activates ATR-ATRIP and promotes TopBP1 recruitment to sites of UV-damage. DNA Repair (Amst). 2014; 21:1-11.
- Acevedo J, Yan S, Michael WM. Direct Binding to Replication Protein A (RPA)-coated Single-stranded DNA Allows Recruitment of the ATR Activator TopBP1 to Sites of DNA Damage. J Biol Chem. 2016; 291: 13124-13131.
- Takeishi Y, Ohashi E, Ogawa K, et al. Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells. 2010; 15: 761-771.
- Rappas M, Oliver AW, Pearl LH. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res. 2011; 39: 313-324.
- Ueda S, Takeishi Y, Ohashi E, et al. Two serine phosphorylation sites in the C-terminus of Rad9 are critical for 9-1-1 binding to TopBP1 and activation of the DNA damage checkpoint response in HeLa cells. Genes Cells. 2012; 17: 807-816.
- Furuya K, Poitelea M, Guo L, et al. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004; 18: 1154-1164.
- Puddu F, Granata M, Di Nola L, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008; 28: 4782-4793.
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. 2009; Mol Cell. 36: 743-753.
- Cotta-Ramusino C, McDonald ER 3rd, Hurov K, et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011; 332: 1313-1317.
- Lindsey-Boltz LA, Kemp MG, Capp C, et al. RHINO forms a stoichiometric complex with the 9-1-1 checkpoint clamp and mediates ATR-Chk1 signaling. Cell Cycle. 2015; 14: 99-108.
- Pandita RK, Sharma GG, Laszlo A, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006; 26: 1850-1864.
- Tsai FL, Kai M. The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle. 2014; 13: 3460-3464.
- Wang Q, Goldstein M, Alexander P, et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014; 33: 862-877.
- Shiotani B, Nguyen HD, Håkansson P, et al. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 2013; 3: 1651-1662.
- Kobayashi M, Hayashi N, Takata M, et al. NBS1 directly activates ATR independently of MRE11 and TOPBP1. Genes Cells. 2013; 18: 238-246.
- Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog TLK1 binds and phosphorylates Rad9 TLK1 acts as a molecular chaperone in DNA repair. DNA Repair (Amst). 2009; 8: 87-102.
- Canfield C, Rains J, De Benedetti A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC Mol Biol. 2009; 10: 110.
- Kelly R, Davey SK. Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit. PLoS One. 2013; 8: e85859.
- Furuya K, Miyabe I, Tsutsui Y, et al. DDK phosphorylates checkpoint clamp component Rad9 and promotes its release from damaged chromatin. Mol Cell. 2010; 40: 606-618.
- Hirai I, Wang HG. A role of the C-terminal region of human Rad9 (hRad9) in nuclear transport of the hRad9 checkpoint complex. J Biol Chem. 2002; 277: 25722-25727.
- Lee MW, Hirai I, Wang HG. Caspase-3-mediated cleavage of Rad9 during apoptosis. Oncogene. 2003; 22: 6340–6346.
- Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J Mol Biol. 2009; 390: 490-502.
- Takeishi Y, Iwaya-Omi R, Ohashi E, et al. Intramolecular Binding of the Rad9 C Terminus in the Checkpoint Clamp Rad9-Hus1-Rad1 Is Closely Linked with Its DNA Binding. J Biol Chem. 2015; 290: 19923-19932.
- Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998; 20: 195-199.
- Ohashi E, Hanafusa T, Kamei K, et al. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009; 14: 101-111.
- Gellon L, Werner M, Boiteux S. Ntg2p a Saccharomyces cerevisiae DNA N-glycosylase/apurinic or apyrimidinic lyase involved in base excision repair of oxidative DNA damage interacts with the DNA mismatch repair protein Mlh1p. Identification of a Mlh1p binding motif. J Biol Chem. 2002; 277: 29963-29972.
- Doré AS, Kilkenny ML, Rzechorzek NJ, et al. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell. 2009; 34: 735-745.
- Eichinger CS, Jentsch S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc Natl Acad Sci U S A. 2010; 107: 11370-11375.
- Chen MJ, Lin YT, Lieberman HB, et al. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J Biol Chem. 2001; 276: 16580-16586.
